Battling Lipid Annotation
March 7th, 2022, by Thao Nguyen
BATL is assembled on our Computational Lipidomics and Metabolomics (CompLiMet) platform
.png)
BATL is assembled on our Computational Lipidomics and Metabolomics (CompLiMet) platform
.png)
Lipidomics is the attempt to map and quantify lipid species sets within a cell or tissue to identify biomarkers and elucidate metabolism at the cellular level. Due to the extensive chemical diversity of lipid species, it is a challenge to measure the lipidome comprehensively and in a single experiment.
Targeted lipidomics utilizing mass spectrometry is a quantitative approach which takes information from discovery experiments and clinical observations to test a hypothesis. This involves verification and validation of defined groups of known lipids across large sample sets. These experiments require accuracy, high throughput and reliability.
For routine quantitation of well-characterized lipids or low-level signaling lipids, selective/multiple reaction monitoring mass spectrometry (SRM/MRM-MS) experiment using triple quadrupole MS is the gold standard. The advantages of SRM include reduced interference, lower detection levels and faster method creation and data acquisition.
In SRM/MRM-MS experiments, signal corresponding to a specific lipid is manually selected, integrated and assigned identities under the assumption that an observed signal within a given retention time corresponds to a single lipid target. Although this is a valid assumption for a single condition, peak features are fundamentally influenced by treatment, disease state, and tissue matrix. Manual curation becomes unreliable when multiple isobars, varying across conditions, are detected within the same transition and retention time window. The human curation approach is thus prone to identification error and biased towards reporting lipids of interest expected by the analyst.
To address these issues, we developed a bioinformatics tool, named Bayesian Annotations for Targeted Lipidomics (BATL), a Gaussian naïve Bayes classifier, which models distributions of input features across biological conditions. BATL annotates peak identities according to eight features related to retention time, intensity, and peak shape. BATL is a useful tool for accurate, targeted lipid identification and, with online access, is easily integrated into any lipidomic pipeline. BATL is assessable on our Computational Lipidomics and Metabolomics (CompLiMet) platform at https://complimet.ca/batl/ .
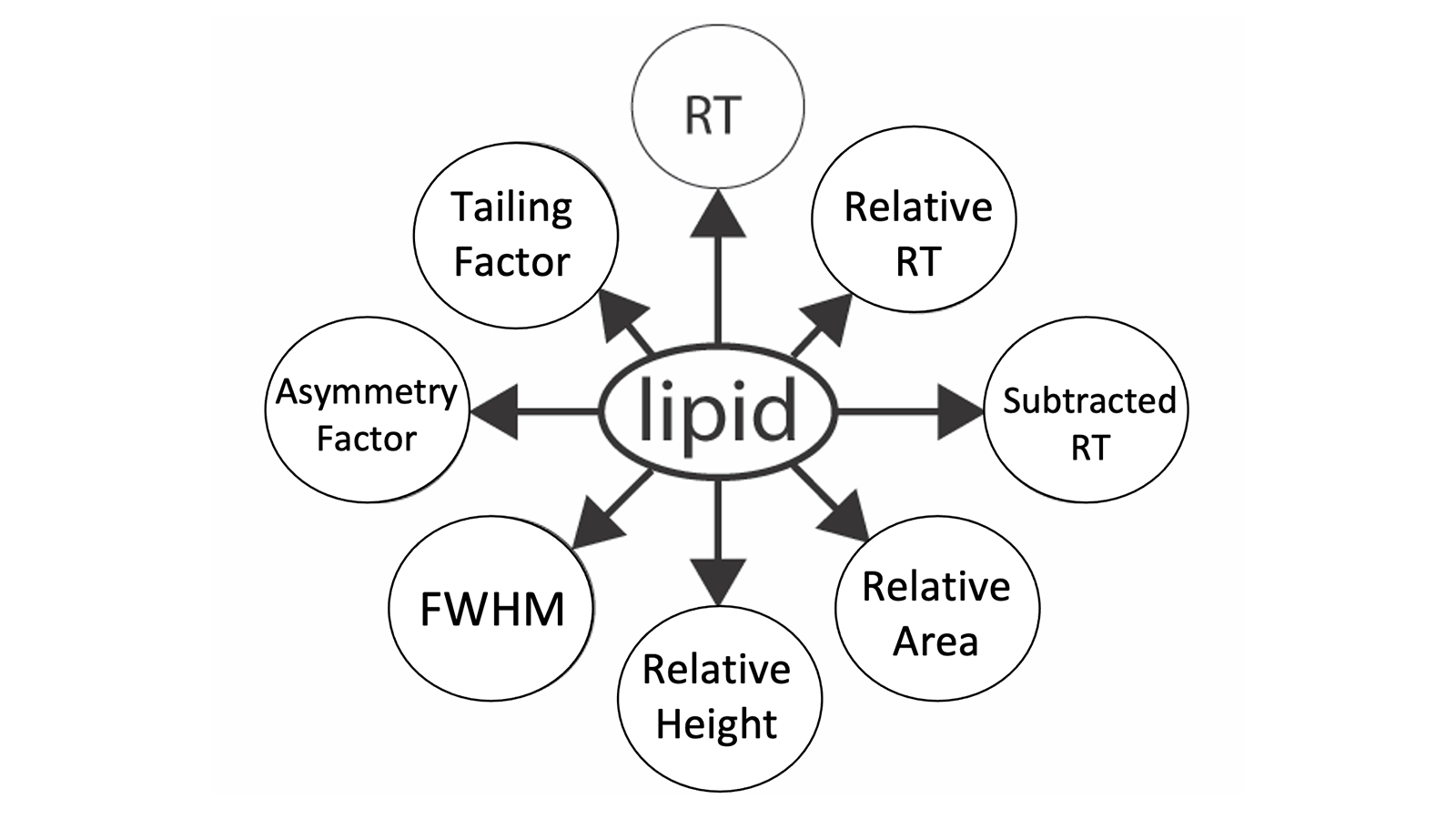
Figure 1 shows the common challenges associated with SRM, MRM and PRM peak identification which BATL seeks to address, while figure 2 displays the eight SRM peak features used for building the naïve Bayes model.