Taking Aim: Identifying potential therapeutic targets against SARS-CoV-2 infection
March 18th, 2022, by Emily Hashimoto-Roth
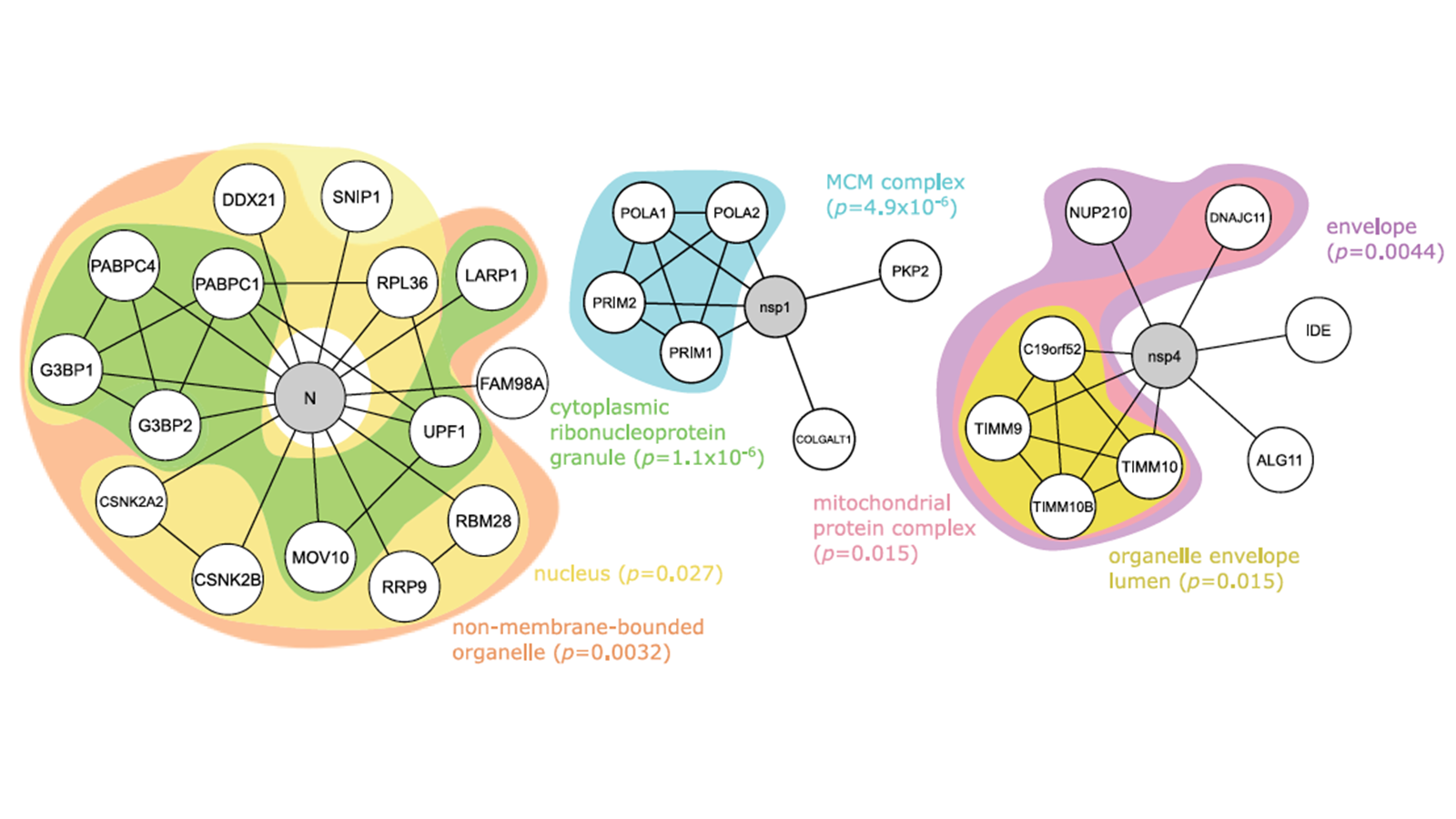

As the COVID-19 pandemic, driven by the SARS-CoV-2 virus, continues, scientists around the globe have collectively come together to develop potential therapeutics against the virus, in addition to the currently available vaccines. The identification of therapeutic targets is naturally a crucial step in this process. One such approach for this task is through the use of protein-protein interaction networks or, more specifically, by computationally mining the protein-protein interactions taking place between viral and host proteins.
In this study, we performed a sophisticated computational analysis of the protein-protein interactions between various SARS-CoV-2 viral proteins and human host proteins, identified and published by Dr. Nevan Krogran’s research group at the University of California, San Francisco, USA (Nature 2020, 583, 459–468). Our analysis began by taking the virus-host protein-protein interaction networks and searching for enriched amino acid sequence motifs, using the Multiple EM for Motif Elicitation (MEME) tool. This analysis was driven by the hypothesis that a significantly enriched sequence motif between viral and host proteins may serve as potential therapeutic targets. To supplement the identification of sequence motifs, we also analyzed the protein-protein interaction networks for significantly enriched functional annotations, using Ontologizer, to identify the biological processes implicated in the protein-protein interactions. Likewise, these significantly enriched biological processes could lead to potential therapeutic targets.
The identification of enriched amino acid sequence motifs and functional annotations became the basis for our further analysis of the viral-host protein-protein interaction networks, as we augmented and extended these networks with known interactions using the STRING database. These notably larger networks were additionally analyzed using LESMoN-Pro, for enriched sequence motifs, and using GoNet, for enriched functional annotations. LESMoN-Pro identified 9 significantly enriched amino acid motifs and GoNet identified 329 Gene Ontology terms, identified to be significantly clustered in the in networks.
Our computational analysis of viral-host protein-protein interaction networks for the identification of enriched motifs and functional annotations implicated with SARS-CoV-2 infection in humans may be useful to future research, providing new potential therapeutic targets.